Modeling Biological Networks
IV.1 Coordinators
IV.2 Participants
IV.3 Introduction
IV.4 Background and Significance
IV.5 Research Plan IV.6 Specific Subprojects
IV.7 Connection to Specific Projects 2 (cytoskeleton) and 3 (organogenesis)
IV.8 Timeline
< Previous | Page 4 of 35 | Next >
IV.5.ii Preliminary Results:
The Consortium will focus initially on molecular level networks. Biological networks control important processes necessary to life at all scales from the molecular level to the level of the complete organism. At the molecular scale, genetic switching, reaction kinetics, and metabolic and gene control networks determine cell function; at the subcellular scale, cytoskeletal networks control cell shape and motion, and signaling networks control most aspects of the cell's proper functioning; at the scale of individual cells, tight junctions transmit signals locally, while nerve fibers can traverse the whole organism; at the tissue level, networks such as heart muscle, lymph nodes, and central pattern generators coordinate vital functions; while at the organ level, the brain, and the hormonal and immune systems regulate large scale function and behavior. Once sufficient resources are available, we intend to expand the work of the Consortium in these directions, since many of the techniques that we will develoop at the molecular level will remain useful at larger scales.
IV.5.ii.a The Topology of Cellular Networks:
The robustness of cellular processes arises from the dynamic interactions among their many constituents (Barkai and Leibler, 1997; Yi et al., 2000; Bhalla and Iyengar, 1999), including proteins, DNA, RNA, and small molecules. Researchers have confirmed the existence of complex interactions among various components of cells and simple microorganisms, but the absence of large-scale databases and a sufficiently developed theoretical framework prevented meaningful analysis of these interactions. Recent large-scale sequencing, coupled with systematic two-hybrid analyses, has now provided complete sequence information for numerous genomes and proteomes (Uetz et al., 2000; Rain et al., 2001) and integrated pathway-genome databases (Overbeek, 2000; Karp et al., 1999; Kanehisa and Goto, 2000) that provide organism-specific connectivity maps of metabolic and, to a lesser extent, other cellular networks. Due to the quantity and diversity of their constituents and reactions, these maps are extremely complex, offering only limited insight into their organizational principles.
Recent advances in understanding the generic properties of complex networks have improved our ability to address the structure of cellular networks quantitatively (Barabasi and Albert, 1999; Albert, Jeong and Barabasi, 2000; Strogatz, 2001).
Until recently, complex network models used classical random network theory (Erdos and Renyi, 1960; Bollobas, 1985), which assumes that each pair of nodes (i.e., constituents) in the network connects randomly with probability p. This process leads to a statistically homogeneous network in which most nodes have approximately the same number of links, k (Figure IV.1 a, b). On the other hand, recent empirical studies have demonstrated that the World-Wide Web (Albert et al., 1999), Internet (Faloutsos et al., 1999), and social networks (Barabasi and Albert, 1999) are scale-free networks (Barabasi and Albert, 1999) (Figure IV.1c), in which P(k) follows a power-law, i.e., P(k) ~ kγ (Figure IV.1 d). Unlike exponential networks, scale-free networks are extremely heterogeneous; a few highly connected nodes (hubs) dominate their topology, linking the remaining, less connected nodes to the network (Figure IV.1 c).
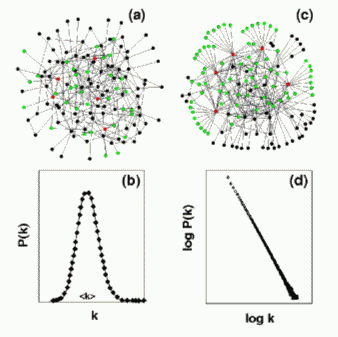
Fig. IV.1. Examples of (a) an exponential and (c) a scale-free network. Connectivity distribution P(k) of (b) an exponential and (d) a scale-free network, giving the probability that a given node connects to k other nodes.
This well-developed theoretical framework, combined with detailed biochemical databases, allows us to begin to analyze cellular networks. Our first questions include: What is the topological structure of metabolic and other cellular networks? What biologically and topologically relevant quantities characterize them? What structural characteristics are generic to all prokaryotic and eukaryotic cells? The following section summarizes our results on the large-scale structure of biochemical reaction pathways (including metabolic- and information-transfer networks), and protein interaction networks.