Modeling Biological Networks
IV.1 Coordinators
IV.2 Participants
IV.3 Introduction
IV.4 Background and Significance
IV.5 Research Plan
IV.6 Specific Subprojects IV.7 Connection to Specific Projects 2 (cytoskeleton) and 3 (organogenesis)
IV.8 Timeline
< Previous | Page 18 of 35 | Next >
IV.6.iii.b Methodology:
We seek to use this clustering to identify regulatory information in the genome.
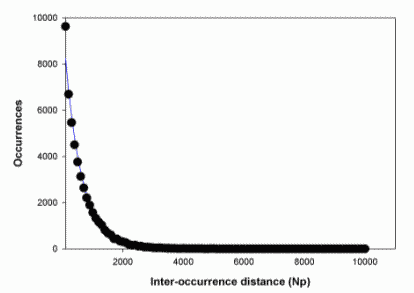
Step 1: Catalog all occurrences of every word of length [4, 12].Step 2: For each word, examine the inter-instance distances and compare to expectation based on a uniform distribution. At present, we do not consider base composition but simply calculate the expected distribution given overall occurrences. We look for words uniformly distributed at large spacings (provisionally, >1000 Np) but clustered at smaller spacings. Figure IV.7 shows that 5'-TCAGT-3' is such a word. The curve shows the expected uniform distribution of the 50,119 instances of this word in the 27,890,790 Np of Drosophila chromosome 3R. The symbols show the good agreement of the data except for an excess of about 1400 instances (ca. 17%) in the distance range [1,100]. Our hypothesis is that words that show this excess are biologically interesting, though we do not assert the inverse. Words that cluster locally may fail this test because of long-range interactions or other non-uniformity.
|
Fig. IV.7. Frequency of distances between known clusters of the sequence 5'-TCAGT-3'. |
Step 3: Look at inter-word distributions. Given words A and B, each of which passes the nonuniformity test, examine the distribution of A-B distances. If the number of candidates is too large, we will look at the most remarkable candidates first and use biological information and intuition.
We will use these statistical properties to identify candidate important words (e.g. the spacings of 5'-TCAGT-3' identify the previously unknown word 5'-TCAGTCAGT-3' as an excellent candidate).
Step 4: Correlate the positions of clustered words to identify "clusters" of likely significance and correlate these with annotated genes.