Modeling Biological Networks
IV.1 Coordinators
IV.2 Participants
IV.3 Introduction
IV.4 Background and Significance
IV.5 Research Plan
IV.6 Specific Subprojects
- IV.6.iv Subproject 4 Modeling the Regulation and Function of Metabolism
- [ Other Subprojects ]
IV.8 Timeline
< Previous | Page 20 of 35 | Next >
IV.6.iv Subproject 4 - Modeling the Regulation and Function of Metabolism:
The data and approaches of Subprojects 1 - 3 present a partial snapshot of metabolism. The topology of the metabolic network describes only genome-encoded potential metabolic activity. The metabolic network, however, actually functions through the genetic regulatory network that activates and inactivates various enzymes or groups of enzymes that catalyze biochemical reactions in the metabolic network. Thus, an in-depth characterization of metabolism requires an understanding of the regulatory network as well.
The goal of Subproject 4 is to go beyond topological characterization to evaluate dynamical correlations between metabolic network pathways. An important limitation of any modeling effort is the lack of availability of enzyme kinetic data, making full dynamic characterization impossible. However, available microarray data provide important qualitative information on correlations between the enzymatic activity of different pathways.
IV.6.iv.a Datasources:For the E. coli transcriptome, microarray data (Arfin et al., 2000; Khodursky et al., 2000; Selinger et al., 2000; Zimmer et al., 2000; Wei et al., 2001) are currently available, and the amount of E. coli transcriptome data should substantially increase during the next five years. We plan to experimentally generate a large amount of E. coli transcriptome data. For corollary analyses in the yeast, S. cerevisiae, we will use data from the Stanford Microarray Database (http://genome-www4.stanford.edu/MicroArray/SMD) and those deposited by Friend (Hughes et al., 2000).
FIG. IV.8. Flowchart of analysis methodology.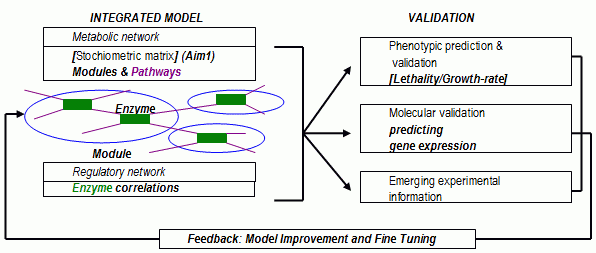
IV.6.iv.b Methods:
1. We will use standard clustering methods to uncover correlations between enzyme concentrations on the transcriptome level.
2. To obtain biological insight into the origin of the correlations, we will use available information on the genetic regulatory organization of our model organisms.
3. Finally, we will use singular value decomposition to uncover the functional limitations of the metabolism.
The outcome of this investigation will be a detailed understanding of cross-correlations between enzymatic activities, as well as a more nuanced and detailed picture of modularity (Subproject 1).