Cytoskeleton and Cell Motility
V.1 Coordinators
V.2 Participants
V.3 Introduction
V.4 Specific Research Objectives
V.5 Background and Significance
V.6 Research Plan
- V.6.i Subproject 1 - Microtubule Plus-end Interacting Proteins
V.8 Timeline
< Previous | Page 5 of 23 | Next >
V.6 RESEARCH PLAN:
V.6.i Subproject 1 - Microtubule Plus-end Interacting Proteins
V.6.i.a GoalsWhat controls the dynamic polymerization of microtubules? How do interactions between polymerizing microtubules and microtubule-binding proteins create complex cytoskeletal structures and functions? Using purified proteins, we can readily recapitulate the dynamic interactions between microtubules and their regulatory proteins in vitro to provide the experimental basis for mathematical models of basic microtubule dynamics and more complex processes such as spindle assembly. If we can model how changes in the concentration or activity of a given regulatory protein affect the overall dynamics of microtubules and then test the model experimentally, we will be able to predict what will happen under conditions that we cannot produce in vitro. We will focus our efforts on proteins (CLIP-170 and XKCM1) that localize and/or act at the plus ends of growing microtubules (Diamantopoulos et al., 1999; Walczak et al., 1996), because these proteins help regulate dynamic instability in vivo (Figure V.3; Brunner and Nurse, 2000; Kline-Smith and Walczak, 2002). While CLIP-170 and XKCM1 are interesting both in themselves and as a test case - CLIP-170 suppresses microtubule depolymerization while XKCM1 promotes it (Brunner and Nurse, 2000; Desai et al., 1999; Diamantopoulos et al., 1999) - the models we develop should apply to additional microtubule binding proteins.
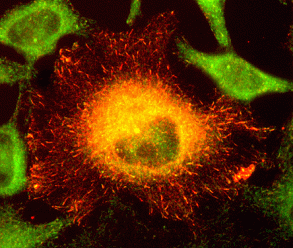 |
Fig. V.3. Localization of transfected CLIP-170 (red) in fixed HeLa cells. |
V.6.i.a.1 Model the effect of individual microtubule binding proteins on microtubule dynamic instability
We will incorporate into previously established models (Chen and Hill, 1985; Davis et al., 2002; Flyvbjerg et al., 1996; Grego et al., 2001; Martin et al., 1993) quantitative interaction parameters for CLIP-170 and XKCM1. We will also develop novel models of microtubule dynamic instability that will serve as foundations for models of more complex cytoskeletal processes.
V.6.i.a.2 Investigate the effect of mixed microtubule binding proteins (CLIP-170 and XCKM1) on microtubule dynamics by iterative cycles of experiment and modelingCLIP-170 promotes processive microtubule growth, while XKCM1 induces catastrophe. We will test experimentally the effects that mixtures of these proteins have on microtubule dynamics. We will use the models derived from individual protein effects to predict the outcomes of various mixtures. The results will tune both models and advance our understanding of microtubule dynamics in vitro and in vivo.