Cytoskeleton and Cell Motility
V.1 Coordinators
V.2 Participants
V.3 Introduction
V.4 Specific Research Objectives
V.5 Background and Significance
V.6 Research Plan
- V.6.iv Subproject 4 - Experiments on the Actin Cytoskeleton
- V.6.iv.a Introduction
- V.6.iv.b Background and Significance
- V.6.iv.c Aims
- V.6.iv.d Biochemical Assembly of F-actin: Thermodynamics
- V.6.iv.e Construction of a Composite Actin Network to Mimic Cytoskeletal Mechanics
V.8 Timeline
< Previous | Page 14 of 23 | Next >
V.6.iv Subproject 4 - Experiments on the Actin Cytoskeleton
V.6.iv.a IntroductionThe dynamic and mechanical properties of the actin cytoskeleton endow the cell with ability to move, change shape, resist stress, and respond to extracellular signals. These properties emerge from complex interactions between polymerizing actin filaments, free actin subunits, actin monomer/polymer binding proteins, and the signal transduction apparatus that regulates these proteins. To understand the properties of the actin cytoskeleton in a quantitative and predictive way, we must identify the proteins that modulate actin polymerization and experimentally characterize their interactions with actin and their effects on actin polymerization.
V.6.iv.b Background and Significance:A great number of actin modulating proteins are known and at least partially characterized. Their activities range from monomer sequestering proteins to nucleation factors (Figure V.8). Recent studies have demonstrated the central role of the arp2/3 complex in forming a dendritic actin network both at the base of the comet tail of Listeria and at the leading edge of several types of animal cells (Figure V.8) (Rohatgi et al., 1999; Pollard et al., 2000). The Wiskott-Aldrich Syndrome protein (WASp) acts as a co-factor for the nucleation of a branched actin network by binding to an arp2/3 complex at the base of each branch. Phosphatidylinositol 4,5 bisphosphate (PIP(2)) and the GTP binding protein CDC42 activate WASp (Egile et al., 1999; Higgs and Pollard, 2000), relaying transmembrane signals from the inner leaflet of the plasma membrane to the cytosol. Because we know the actin interactions of many of these proteins quantitatively, we can imagine a comprehensive model of the actin cytoskeleton.
However, the mechanisms are far from complete. Does the branched actin network physically attach to the membrane? If so (Loisel et al., 1999; Kuo and McGrath, 2000), how does it generate force? What are the roles of other proteins that either enhance or inhibit actin dynamics at the leading edge? How do interactions between these and other proteins lead to the observed physical properties of the actin cytoskeleton? Have we identified all the relevant actin-modulating proteins?
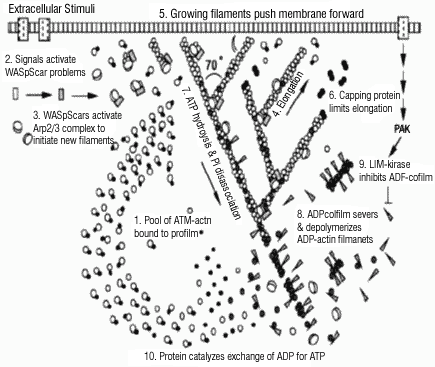 |
| Fig. V.8. The dendritic nucleation model for actin assembly at the leading edge of a motile cell (from Pollard et al., 2000). |
V.6.iv.c Aims:
We propose the following Specific Subprojects:
1. Biochemical Assembly of F-actin: Thermodynamics.
2. Construction of a composite actin network to mimic the cytoskeletal mechanics.
3. Regulatory Alteration of Actin Binding Proteins.
4. Computational Modeling of the Actin Cytoskeleton.