Cloud Technologies for Bioinformatics Applications
Abstract
Test the performance variation of couple of Bioinformatics applications running on Apache Hadoop on Linux Virtual Machines and on Microsoft DryadLINQ on Windows HPCS cluster over multiple runs.
Intellectual Merit
Analyzing the performance and viability of cloud technologies to conduct bioinformatics data analyses.
Broader Impact
We analyze the feasibility of cloud environments to conduct bioinformatics data analyses, providing recommendations and guidance for the bio-informatics scientists.
Use of FutureGrid
We are going to test the performance variation of couple of Bioinformatics applications running on Apache Hadoop on Linux Virtual Machines and on Microsoft DryadLINQ on Windows HPCS cluster over multiple runs.
Scale Of Use
"33 Nodes (8 cores per node) on the Windows HPCS 2008 cluster.33 Nodes (8 cores per node) Xen VM cluster with the instances running Linux with access to local disks and a shared file system."
Publications
Results
Ongoing with results:
For the first step of our project, we performed an in-detail performance analysis of different implementations of two popular bio-informatics applications, namely sequence alignment using SmithWaterman-GOTOH algorithm and sequence assembly using CAP3 program. These applications were implemented using cloud technologies such as Hadoop MapReduce and Microsoft DryadLINQ as well as using MPI. The performance comparison consisted of comparing the performance scalability of the different implementations, analyzing the effects of inhomogeneous data on the performance of cloud technology implementations and comparing the performance of cloud technology implementations under virtual and non-virtual (bare metal) environments. We also performed an auxiliary experiment to calculate the systematic error of these applications in different environments.
We used Apache Hadoop on 33 bare metal Linux Futuregrid nodes as well as on 33 future grid Linux virtual instances (deployed using Eucalyptus). We also used Microsoft DryadLINQ on 33 bare metal Windows HPCS cluster on Futuregrid.The results are published in the following paper.
Following graphs present few selected results from our project. For more information refer to the above paper.
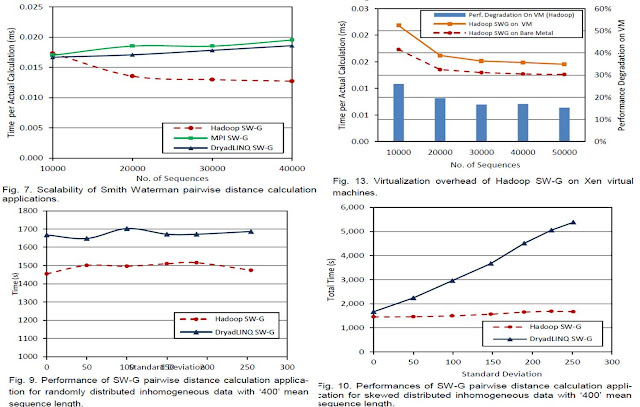
For the second step of our project, we implemented few pleasingly parallel bio-medical applications using cloud technologies, Apache Hadoop MapReduce and Microsoft DryadLINQ, and using cloud infrastructure services provided by commercial cloud service providers, naming it the "Classic Cloud" model. The applications used were sequence assembly using Cap3, sequence alignment using BLAST, Generative Topographic Mapping (GTM) interpolation and Multi Dimensional Scaling (MDS) interpolation. We used Amazon EC2 and Microsoft Windows Azure platforms for obtaining the "Classic Cloud" implementation performance results, while we used FutureGrid compute resources to obtain the Apache Hadoop and Microsoft DryadLINQ performance results. The results were published in the following papers.
Thilina Gunarathne, Tak-Lon Wu, Judy Qiu, and Geoffrey Fox, Cloud Computing Paradigms for Pleasingly Parallel Biomedical Applications March 21 2010. Proceedings of Emerging Computational Methods for the Life Sciences Workshop of ACM HPDC2010 conference, Chicago, Illinois, June 20-25, 2010.
Thilina Gunarathne, Tak-Lon Wu, Jong Youl Choi, Seung-Hee Bae, Judy Qiu Cloud Computing Paradigms for Pleasingly Parallel Biomedical Applications, Submitted for publication in ECMLS special edition of Concurrency and Computations Journal (invited).


 Futuregrid is a resource provider for
Futuregrid is a resource provider for