FG-18
Contemporary {HPC} Architectures
Privacy preserving gene read mapping using hybrid cloud
Project Details
- Project Lead
- Yangyi Chen
- Project Manager
- Yangyi Chen
- Project Members
- KEHUAN ZHANG
- Supporting Experts
- Javier Diaz Montes, Andrew Younge
- Institution
- Indiana University Bloomington, School of Informatics and Computing
- Discipline
- Computer Science (401)
Abstract
We would like to study the possibility of doing reads mapping using hybrid cloud, in order to utilize public computing resources while preserving the data privacy.
Intellectual Merit
This research if of high demand in the area of bioinformatics as more and data are generated everyday but lack of computing resources to process them.
Broader Impacts
The research may increase data processing speed in the area of bioinformatics and thus replace current read mapping tools
Scale of Use
Run experiments on the system and for each experiment I will need about 2~3 days.
Results
One of the most important analyses on human DNA sequences is read mapping, which aligns a large number of short DNA sequences (called reads) produced by sequencers to a reference human genome. The analysis involves intensive computation (calculating edit distances over millions upon billions of sequences) and therefore needs to be outsourced to low-cost commercial clouds. This asks for scalable privacy-preserving techniques to protect the sensitive information sequencing reads contain. Such a demand cannot be met by the existing techniques, which are either too heavyweight to sustain data-intensive computations or vulnerable to re-identification attacks. Our research, however, shows that simple solutions can be found by leveraging the special features of the mapping task, which only cares about small edit distances, and those of the cloud platform, which is designed to perform a large amount of simple, parallelizable computation. We implemented and evaluated such new techniques on a hybrid cloud platforms built on FutureGrid. In our experiments, we utilized specially-designed techniques based on the classic “seed-and-extend” method to achieve secure and scalable read mapping. The high-level design of our techniques is illustrated in the following figure: the public cloud on FutureGrid is delegated the computation over encrypted read datasets, while the private cloud directly works on the data. Our idea is to let the private cloud undertake a small amount of the workload to reduce the complexity of the computation that needs to be performed on the encrypted data, while still having the public cloud shoulder the major portion of a mapping task.
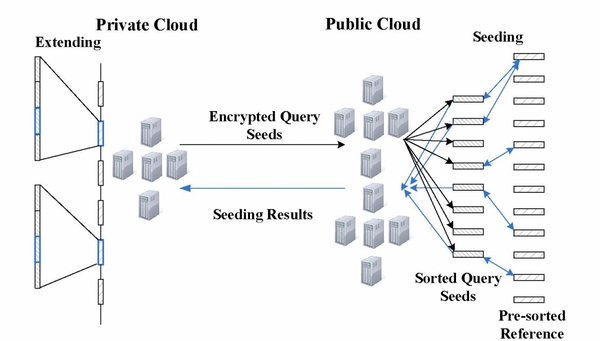
We constructed our hybrid environment over FutureGrid in the following two modes:
1. Virtual mode:
We used 20 nodes on FutureGrid as the public cloud and 1 node as the private cloud.
2. Real mode:
We used nodes on FutureGrid as the public cloud and the computing system within the School of Informatics and Computing as the private cloud. In order to get access to the all the nodes on public cloud, we copied a public SSH key shared by all the private cloud nodes to the authorized_keys files on each public cloud node.
Our experiments demonstrate that our techniques are both secure and scalable. We successfully mapped 10 million real human microbiome reads to the largest human chromosome over this hybrid cloud. The public cloud took about 15 minutes to do the seeding and the private cloud spent about 20 minutes on the extension. Over 96% of computation was securely outsourced to the public cloud.


 Futuregrid is a resource provider for
Futuregrid is a resource provider for